Introduction
Oligonucleotide synthesis provides custom-made oligonucleotides. The purity of oligonucleotide preparations produced by solid-phase synthesis can be very high. However, purification is often required to remove incomplete or erroneous sequences. Anion exchange chromatography (AIEX) is an attractive choice since it can separate the negatively charged oligonucleotides based on length. To obtain a purification that gives high purity and yield of the full-length oligonucleotide, optimization of the binding and elution conditions is needed. We have compared gradient elution conditions for a 20-mer phosphodiester oligonucleotide using NaCl in the presence of 20 mM Tris-HCl, pH 8 or 20 mM NaOH, pH 12 using the agarose-based resin WorkBeads™ 40Q (Bio-Works).
Purification optimization
The resolution of oligonucleotide purifications on WorkBeads 40Q was compared for NaCl-elution in the presence of 20 mM Tris-HCl, pH 8 and 20 mM NaOH, pH 12. The dynamic binding capacity (DBC) was determined by frontal analysis at 150 cm/h to 48 mg/mL resin by applying the crude oligonucleotide preparation from the solid support without further adjustment of the feed.

Column: WorkBeads 40Q (6.6 × 100 mm, 3.4 mL)
Sample: 1 mL of 1 mg/mL crude preparation
from a 20-mer oligonucleotide (ssDNA)
synthesis batch/trityl-OFF
Flow rate: 150 cm/h, 0.86 mL/min
Binding buffers: A. 20 mM NaOH, pH 12
B. 20 mM Tris-HCl, pH 8
Elution buffers: A. 20 mM NaOH, 1.5 M NaCl, pH 12
B. 20 mM Tris-HCl, 1.5 M NaCl, pH 8
Gradients: 1. Step: 20% elution buffer, 10 CV (column volumes)
2. Linear: 20% to 40% or 50%, 20 CV
Figure 1. Purification of oligonucleotides on WorkBeads 40Q using 20 mM NaOH, pH 12 (solid blue) or 20 mM Tris-HCl, pH 8 (dotted green). The elution gradients are shown in red (dotted line 20-40% for 20 mM Tris-HCl, pH 8, and solid line 20-50% for 20 mM NaOH, pH 12).
The main peak obtained was collected in 1-mL fractions. Each individual fraction was analyzed for yield and purity on a DNAPac PA200 analytical IEX column (Thermo Fisher Scientific).

Figure 2. Yield and purity determination. (A) Purification of crude preparation on WorkBeads 40Q. Blue area represents collected main peak. (B) Crude preparation analyzed on DNAPac PA200. (C) Collected main peak fraction analyzed on DNAPac PA200.
Purity and yield
For optimization, selected 1-mL fractions were combined to assess the effect of pooling on yield and purity, see Fig. 3 and Table 1. A purity of 95.5% with 76.9% yield was obtained with the broadest pooling using NaOH buffer (Fig. 3A), and a purity of 95.6% with 74.2% yield using Tris-buffer (Fig. 3B). Higher purities could be obtained by a more narrow pooling.
The WorkBeads 40Q resin was also compared to Capto™ Q ImpRes resin (GE Healthcare). Capto Q ImpRes gave for unknown reason a broader main peak than WorkBeads 40Q (Fig. 3D), despite that the columns showed the same efficiency after packing (data not shown).
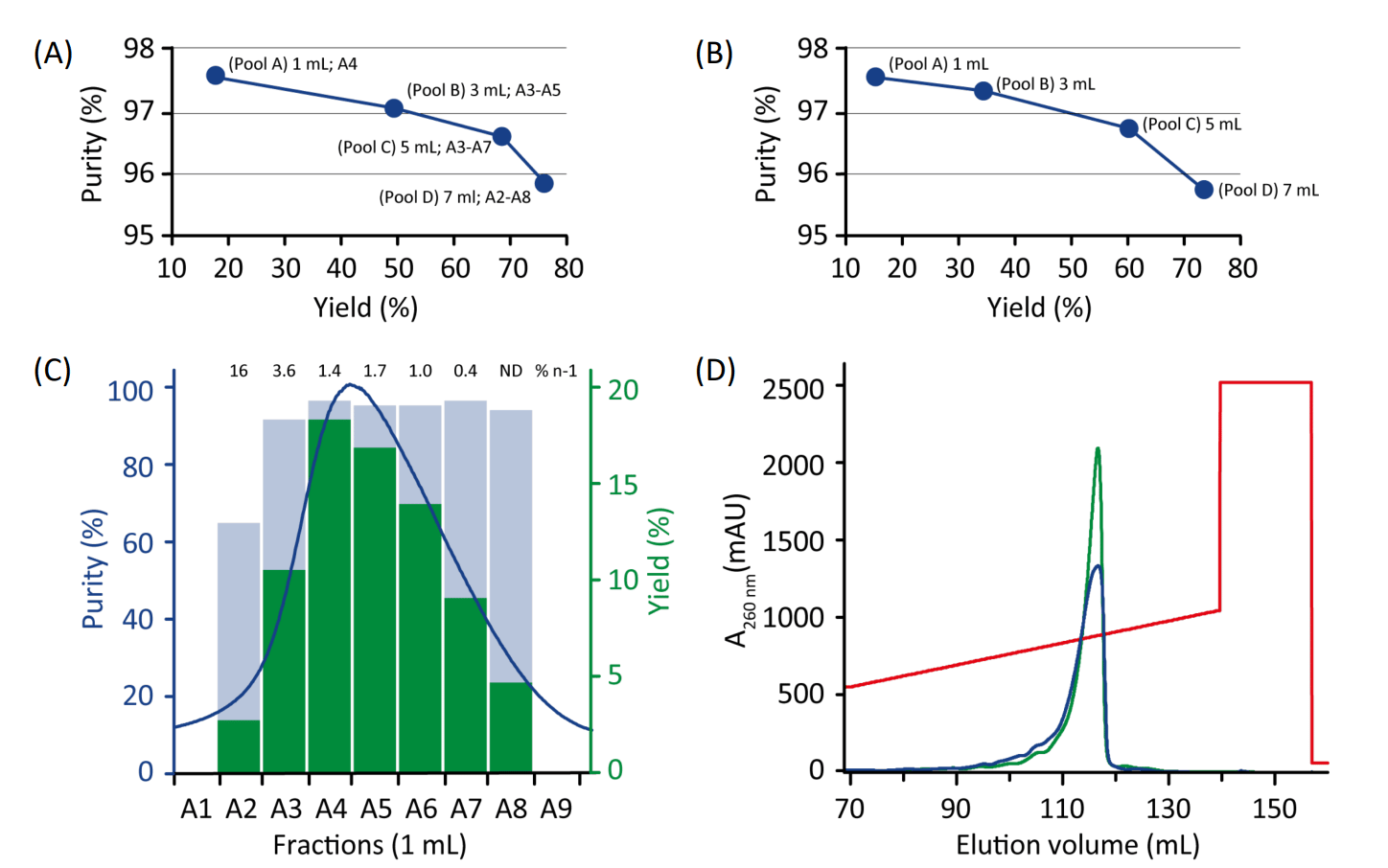
Figure 3. Purity vs yield plots for different fraction pools (1 mL, 3 mL, 5 mL and 7 mL) of the eluted main peak obtained with 20 mM NaOH, pH 12 (A) or, 20 mM Tris-HCl, pH 8 (B). Purity and yield were measured in each individual fraction across the main peak using 20 mM NaOH, pH 12. Contents of n-1 mer are given above the bars (C). Purification on WorkBeads 40Q (green) and Capto Q ImpRes (blue) in 20 mM Tris-HCl, pH 8 (D).
Table 1. Yield and purity. Pools of seven 1 mL fractions of the main peak were compared.

Capto is a trademark of General Electric Company.
Conclusions
Purification of the 20-mer oligonucleotide using WorkBeads 40Q gave excellent purity with good yield in both NaOH- and Tris-based buffers. Both purity and yield were higher on WorkBeads 40Q compared to Capto Q ImpRes, which also gave a broader main peak. The dynamic binding capacity at 150 cm/h was 48 mg/mL resin when applying a crude preparation of oligonucleotide. Trials using NaClO4 as elution salt or addition of organic solvents, e.g, acetonitrile to the buffer systems did not improve the purification (data not shown).
PS40100001 AB